Common oral bacteria can shape their communities in part by their ability to glide over surfaces and transport other types of bacteria that can’t move on their own, according to the National Institutes of Dental and Craniofacial Research (NIDCR). These gliding processes may contribute to the formation of oral biofilms such as plaque. Understanding how these communities form could lead to new ways to prevent or treat a variety of conditions including periodontal disease, reports study author and Harvard postdoctoral researcher Abhishek Shrivastava, PhD.
Shrivastava began his postdoctoral work studying a molecular motor system that enables Flavobacteria, a genus of soil and water microbes, to propel themselves via gliding. He soon learned that an identical motor system was found in a different type of bacteria, Capnocytophaga gingivalis, prevalent in human gingiva and oral biofilms. He then wondered if C gingivalis also moved by gliding and, if so, what role the gliding bacteria play in the oral cavity.
Having never worked in oral microbiology and unsure where to start, Shrivastava contacted Leslie Frieden, PhD, a program officer in NIDCR’s Research Training and Career Development Program. With Frieden’s advice and encouragement, Shrivastava successfully applied for a Pathway to Independence Award, a funding method that helps postdoctoral scholars transition to independent research careers.
Shrivastava found NIDCR-funded oral microbiome researchers at the Forsyth Institute in nearby Cambridge, Massachusetts, who gave him a sample of C gingivalis taken from human periodontal lesions. Shrivastava grew the bacteria and viewed them under the microscope. Just as suspected, he said, “the bacteria were gliding across the slide, very similar to how the Flavobacteria behaved.”
To more thoroughly document the behavior of C gingivalis, Shrivastava and his colleagues spread micron-sized air bubbles across a gelatinous agar surface and added the bacteria, which secreted chemicals that formed a fluid layer. The bacteria then moved the fluid and buoyant air bubbles as they migrated across the agar.
By tracking the bubbles’ positions and movement of the fluid, the researchers could trace the paths of the bacteria. This method showed that C gingivalis bacteria moved together in groups, much like flowing traffic or swarms of bees. The swarms moved in counterclockwise circles and formed layers on top of each other.
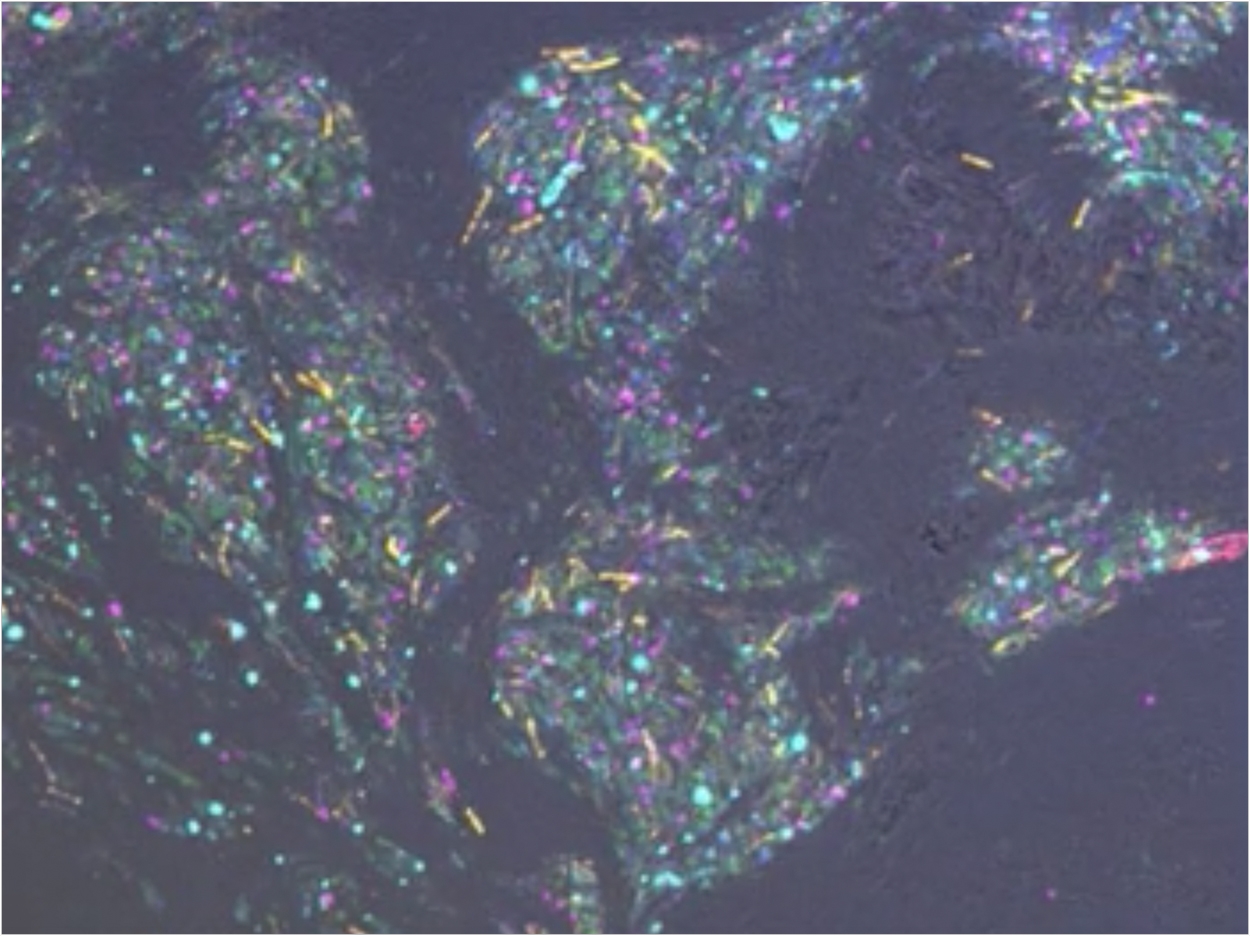
The scientists then designed a series of experiments to more closely replicate how C gingivalis might behave in the oral cavity. They selected seven nonmotile bacterial species that are highly abundant in human gingiva. One at a time, each nonmotile species was placed with C gingivalis on a glass surface. In each case, C gingivalis adhered to and carried the nonmotile bacteria as cargo across the surface.
In another experiment, the researchers labeled each of the nonmotile species with different colored fluorescent tags and placed all eight species together. Again, C gingivalis carried the nonmotile bacteria. Within 20 minutes, the nonmotile bacteria clumped together in islands around which moved swarms of C gingivalis.
“Our findings suggest that gliding bacteria like C gingivalis have evolved a balance of motile and adhesive forces,” said Shrivastava. “These forces drive surface motility and cargo transport, by which the bacteria are able to shape specific structures.
These processes may underlie the formation of oral biofilms, the researchers suggest.
As he finishes his postdoctoral work, Shrivastava anticipates transitioning to his own lab and continuing to explore the forces that shape oral bacterial communities. For example, he said, “how does thickness of saliva, stiffness of tissue, or presence of periodontal disease affect the structure of a community?”
With this information, Shrivastava said, it might be possible to shift the oral microenvironment from a diseased state to a healthier one. Shrivastava also credits NIDCR with helping him push outside his comfort zone.
“I was driven by interesting scientific questions to move beyond my training, and I’m thankful for the opportunity to take some risks that led to a whole new line of inquiry,” he said.
Related Articles
F Nucleatum Accelerates Colon Cancer Growth
Grant to Explore Strategies for Genetically Engineering Oral Microbes
More Than 80 Species Added to Oral Microbiome Database












